CilioGenics
This project consists of integrating protein interaction, comparative genomics, RNA-seq, data mining and motif data to find novel candidate ciliary genes.
This page contains links to some of my projects that showcase my competences in bioinformatics. The projects are grouped in:
The following projects span a wide variety of bioinformatics topics, and are available on github.
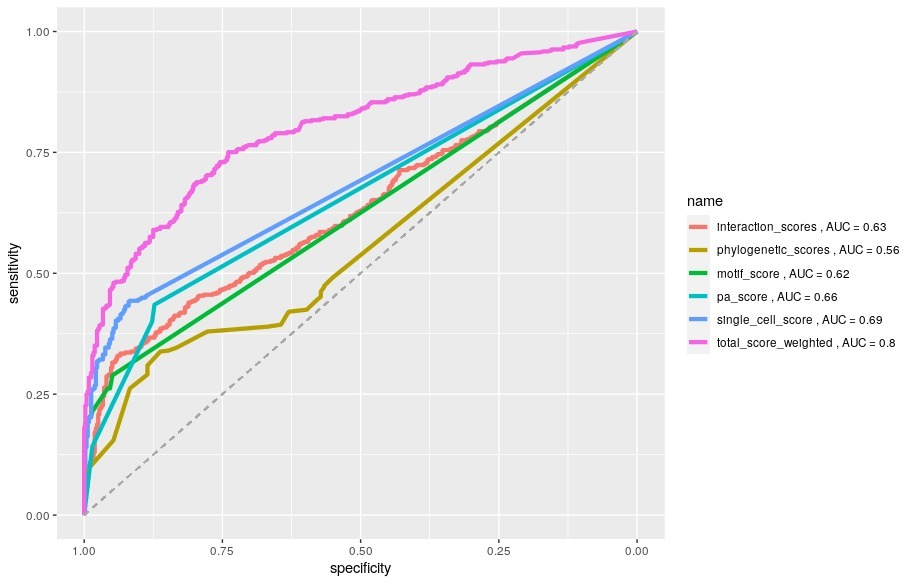
This project consists of integrating protein interaction, comparative genomics, RNA-seq, data mining and motif data to find novel candidate ciliary genes.

ConVarT is a search engine used to match human genetic variants with variants from non-human species. This repo contains pipeline to generate variant data and find matching variants between the species human, mouse and C. elegans. For more information, click here.
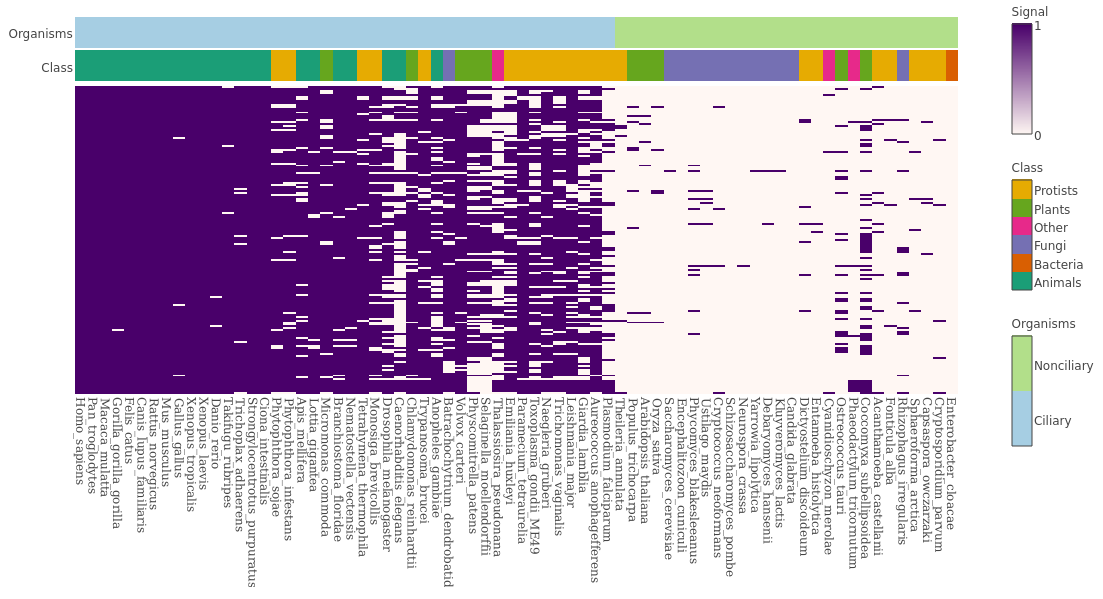
This project explores the genes related to ciliogenesis through comparing genomes of 72 different organisms. It first blasts through genomic sequences to find orthologous genes, then clusters genes by generating phylogenetic tree via hierarchical clustering. Both bash and R scripts are used.
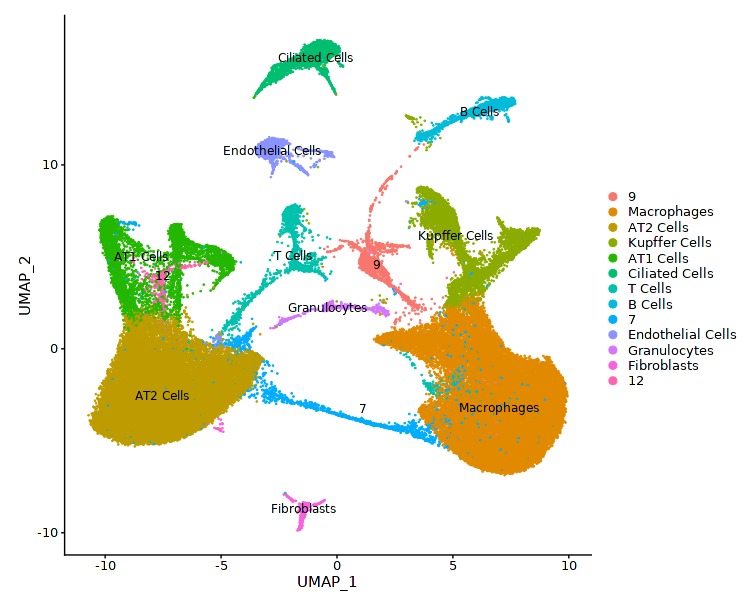
This notebook analyzes publicly available raw lung single cell RNA-seq data ( GSE167295 ) to find genes differentially expressed in ciliated cells.
The following projects span a wide variety of R packages and shiny apps.
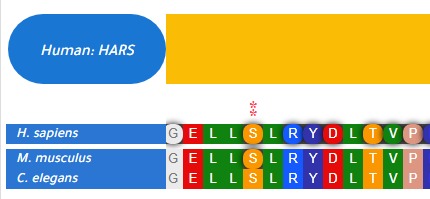
This R package is created to find matching amino acid variants between orthologous protein among different organisms. orthoMSA function is used to generate multiple sequence alignments by only providing organism names as input. Then orthoFind function can be used to find matching variants using generated msa table and user provided variant data.
This shiny app is created to explore the data generated in CilioGenics project.